Likelihood profiling is a key model diagnostic that helps identify the influence of information sources on model estimates. The number of recruits in an unfished population (R0) is a commonly profiled parameter since it dictates the absolute size of a fish population (serves as global scaling parameter). To conduct a profile, values of the parameter over a given range are fixed and the model re-run and then changes in total likelihood and data-component likelihoods are examined. It is recommended to run this throughout the development process, particularly after incorporating new data sets to understand how informative each component is on the estimation of R0 and if there is conflict between data sources.
Model inputs
To run a Stock Synthesis model, four input files are required:
starter.ss, forecast.ss, control.ss, and data.ss. The input files for
the example model can be found within the ss3diags package
and accessed as shown below. Also, if you do not have the
r4ss package installed, you will need to install it for
this tutorial.
install.packages("pak")
pak::pkg_install("r4ss/r4ss")
library(r4ss)
files_path <- system.file("extdata", package = "ss3diags")
dir_tmp <- tempdir(check = TRUE)
dir_profile <- file.path(dir_tmp, "profile")
dir.create(dir_profile, showWarnings = FALSE, recursive = TRUE)
list.files(files_path)
#> [1] "control.ss" "data.ss" "forecast.ss" "starter.ss"
file.copy(from = list.files(files_path, full.names = TRUE), to = dir_tmp)
#> [1] TRUE TRUE TRUE TRUEYou will need to make sure you have the SS
executable file either in your path or in the directory you are
running the profile from (in this case dir_profile). An
easy way to get the latest release of stock synthesis is to use the
r4ss function get_ss3_exe().
r4ss::get_ss3_exe(dir = dir_tmp, version = "v3.30.21")
#> The stock synthesis executable for Linux v3.30.21 was downloaded to: /tmp/RtmpMtDQGk/ss3Running a Profile on R0
Once you have the four input files and SS executable file, you can
run a likelihood profile as shown below. The first step is to run the
model you would like to do a profile for. We will do this in
dir_tmp and then copy the necessary files to
dir_profile. It’s best to run the profile in a subdirectory
of your main model run to keep the output files separate from other
diagnostic tests you may run.
r4ss::run(dir = dir_tmp, exe = "ss3", verbose = FALSE)
#> [1] "ran model"
files <- c("data.ss", "control.ss_new", "starter.ss", "forecast.ss", "ss.par", "ss3")
file.copy(from = file.path(dir_tmp, files), to = dir_profile)
#> [1] TRUE TRUE TRUE TRUE TRUE TRUEOnce you have the input files in the dir_profile
directory, you will need to create a vector of values to profile across.
The range and increments to choose depend on your model and the
resolution at which you want to analyze the likelihood profile. For this
example we will use a fairly coarse resolution to speed up total run
time.
CTL <- SS_readctl_3.30(file = file.path(dir_profile, "control.ss_new"), datlist = file.path(dir_profile, "data.ss"))
CTL$SR_parms
#> LO HI INIT PRIOR PR_SD PR_type PHASE env_var&link dev_link
#> SR_LN(R0) 6.0 12 8.9274 10.3 10.0 0 1 0 0
#> SR_BH_steep 0.2 1 0.8000 0.8 1.0 0 -4 0 0
#> SR_sigmaR 0.0 2 0.6000 0.8 0.8 0 -4 0 0
#> SR_regime -5.0 5 0.0000 0.0 1.0 0 -4 0 0
#> SR_autocorr 0.0 0 0.0000 0.0 0.0 0 -99 0 0
#> dev_minyr dev_maxyr dev_PH Block Block_Fxn
#> SR_LN(R0) 0 0 0 0 0
#> SR_BH_steep 0 0 0 0 0
#> SR_sigmaR 0 0 0 0 0
#> SR_regime 0 0 0 0 0
#> SR_autocorr 0 0 0 0 0
# getting the estimated r0 value
r0 <- CTL$SR_parms$INIT[1]
# creating a vector that is +/- 1 unit away from the estimated value in increments of 0.2
r0_vec <- seq(r0 - 1, r0 + 1, by = 0.2)
r0_vec
#> [1] 7.9274 8.1274 8.3274 8.5274 8.7274 8.9274 9.1274 9.3274 9.5274 9.7274
#> [11] 9.9274We also need to modify the starter file by changing the name of the control file that it will read from and making sure the likelihood is calculated for non-estimated quantities.
START <- SS_readstarter(file = file.path(dir_profile, "starter.ss"), verbose = FALSE)
START$prior_like <- 1
START$ctlfile <- "control_modified.ss"
SS_writestarter(START, dir = dir_profile, overwrite = TRUE, verbose = F)To run the profile, use r4ss::profile() and you will
need to specify a partial string of the name of the parameter you are
profiling over (in this case “SR_LN” will match with “SR_LN(R0)”), and
the vector of values to profile across. The newctlfile is
the control file that will be adjusted with values from the
profilevec and can be named anything you prefer, it just
needs to match what you put in the starter file for “ctlfile”. Full
documentation of the profile() function can be found on the
r4ss
website.
profile(
dir = dir_profile,
newctlfile = "control_modified.ss",
string = "SR_LN",
profilevec = r0_vec,
exe = "ss3",
verbose = FALSE
)
#> running profile i=1/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 7.9274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=2/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 8.1274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=3/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 8.3274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=4/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 8.5274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=5/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 8.7274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=6/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 8.9274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=7/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 9.1274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=8/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 9.3274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=9/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 9.5274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=10/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 9.7274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> running profile i=11/11
#> Parameter names in control file matching input vector
#> 'strings' (n=1): SR_LN(R0)
#> line numbers in control file (n=1): 120
#> Wrote new file to control_modified.ss with the following changes:
#> oldvals newvals oldphase newphase oldlos newlos oldhis newhis oldprior
#> 1 8.9274 9.9274 1 -1 6 6 12 12 10.3
#> newprior oldprsd newprsd oldprtype newprtype comment
#> 1 10.3 10 10 0 0 # SR_LN(R0)
#> Value converged TOTAL Catch Equil_catch Survey Length_comp
#> 1 7.9274 TRUE 1067.570 4.40715e-01 16.620400 6.065520 453.198
#> 2 8.1274 TRUE 937.149 1.46579e+00 13.928400 1.888540 417.316
#> 3 8.3274 TRUE 844.325 4.84432e-01 9.292590 -2.134720 393.233
#> 4 8.5274 TRUE 790.513 9.17170e-06 3.930790 -5.726270 379.721
#> 5 8.7274 TRUE 762.562 6.48949e-08 0.271032 -7.140910 370.420
#> 6 8.9274 TRUE 754.302 1.33458e-10 0.174869 -7.118110 366.756
#> 7 9.1274 TRUE 758.927 2.10816e-13 1.337060 -6.106700 367.497
#> 8 9.3274 TRUE 767.708 1.90347e-13 3.424990 -4.917570 369.543
#> 9 9.5274 TRUE 777.243 1.15331e-13 6.433440 -3.857200 371.605
#> 10 9.7274 TRUE 787.189 6.73658e-14 10.382600 -2.563580 373.376
#> 11 9.9274 TRUE 798.152 4.05051e-14 15.279300 -0.380772 374.815
#> Age_comp Size_at_age Recruitment InitEQ_Regime Forecast_Recruitment
#> 1 341.340 230.715 19.181500 0.00000e+00 0
#> 2 289.737 199.683 13.121200 0.00000e+00 0
#> 3 258.805 179.372 5.270210 0.00000e+00 0
#> 4 243.456 167.416 1.713560 1.54502e-30 0
#> 5 234.970 163.525 0.515980 1.54502e-30 0
#> 6 231.845 162.399 0.244060 1.54502e-30 0
#> 7 233.210 162.675 0.312441 0.00000e+00 0
#> 8 235.957 163.170 0.529082 0.00000e+00 0
#> 9 238.622 163.598 0.839498 1.54502e-30 0
#> 10 240.872 163.942 1.178030 0.00000e+00 0
#> 11 242.718 164.208 1.510550 0.00000e+00 0
#> Parm_priors Parm_softbounds Parm_devs Crash_Pen max_grad
#> 1 0 0.00756177 0 0 1.53717e-05
#> 2 0 0.00997359 0 0 4.45599e-05
#> 3 0 0.00312060 0 0 6.84977e-06
#> 4 0 0.00197985 0 0 2.05016e-05
#> 5 0 0.00143663 0 0 5.99653e-06
#> 6 0 0.00141332 0 0 1.69256e-05
#> 7 0 0.00150262 0 0 4.91877e-06
#> 8 0 0.00159065 0 0 7.52242e-06
#> 9 0 0.00166199 0 0 1.63161e-05
#> 10 0 0.00171844 0 0 1.71582e-07
#> 11 0 0.00176282 0 0 2.46224e-05Visualizing Output
Once the profile is finished running, we can visualize the results to determine if there is conflict between the data sources. If all data sources reach a negative log-likelihood (NLL) minimum at the same R0 value, this indicates good agreement between them. However, more likely, one or more will reach this NLL minimum at different R0 values from the global R0 value. This is a sign of conflict between your data sources and may require you to consider data weighting.
profile_mods <- SSgetoutput(dirvec = dir_profile, keyvec = 1:length(r0_vec), verbose = FALSE)
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
#> Warning in SS_output(dir = mydir, repfile = repFileName, covarfile = covarname, : 32 estimated parameters indicated by the par file
#> 31 estimated parameters shown in the covar file
#> Returning the par file value: 32
profile_mods_sum <- SSsummarize(profile_mods, verbose = FALSE)
SSplotProfile(profile_mods_sum,
profile.string = "SR_LN",
profile.label = "SR_LN(R0)"
)
#> Parameter matching profile.string=SR_LN: SR_LN(R0)
#> Parameter values (after subsetting based on input 'models'): 7.9274, 8.1274, 8.3274, 8.5274, 8.7274, 8.9274, 9.1274, 9.3274, 9.5274, 9.7274, 9.9274
#> Likelihood components showing max change as fraction of total change.
#> To change which components are included, change input 'minfraction'.
#> frac_change include label
#> TOTAL 1.0000 TRUE Total
#> Catch 0.0047 FALSE Catch
#> Equil_catch 0.0525 TRUE Equilibrium catch
#> Survey 0.0422 TRUE Index data
#> Length_comp 0.2759 TRUE Length data
#> Age_comp 0.3495 TRUE Age data
#> Size_at_age 0.2181 TRUE Size-at-age data
#> Recruitment 0.0605 TRUE Recruitment
#> InitEQ_Regime 0.0000 FALSE Initital equilibrium recruitment
#> Forecast_Recruitment 0.0000 FALSE Forecast recruitment
#> Parm_priors 0.0000 FALSE Priors
#> Parm_softbounds 0.0000 FALSE Soft bounds
#> Parm_devs 0.0000 FALSE Parameter deviations
#> Crash_Pen 0.0000 FALSE Crash penalty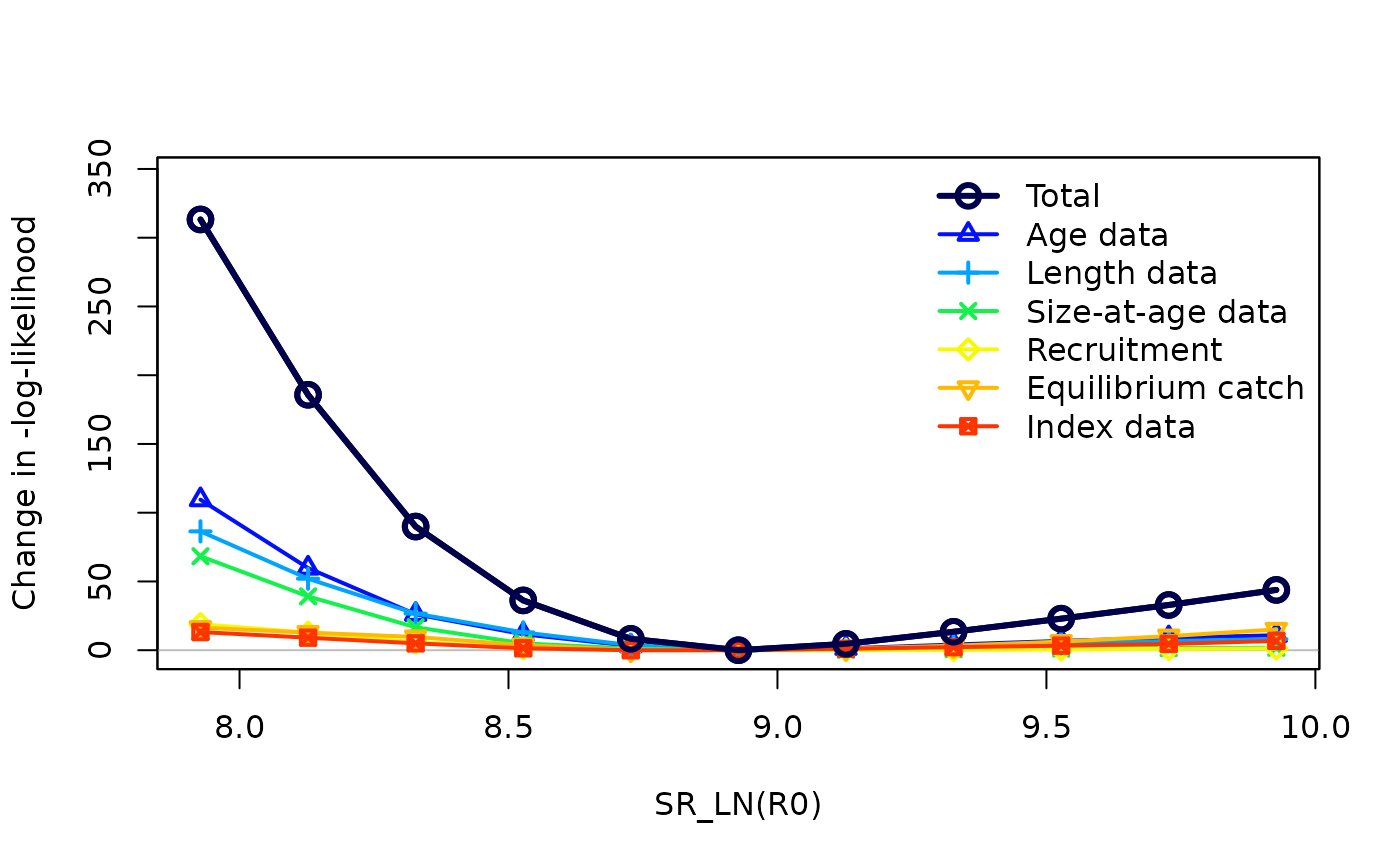
The profile plot above shows the changes in log-likelihood across the selected range of values for each of the contributing data components. There is an adjustable minimum contribution threshold that a component must meet to appear in this figure (if there is a data source that is in your model but does not show up in the plot, its contribution to the total likelihood may not be large enough). The steepness of each trajectory indicates how informative (or not) a data source is. For example, the age data in the plot above is much steeper on the left side of the minimum R0 value than the index data, which suggests that age composition data is more informative in the model.
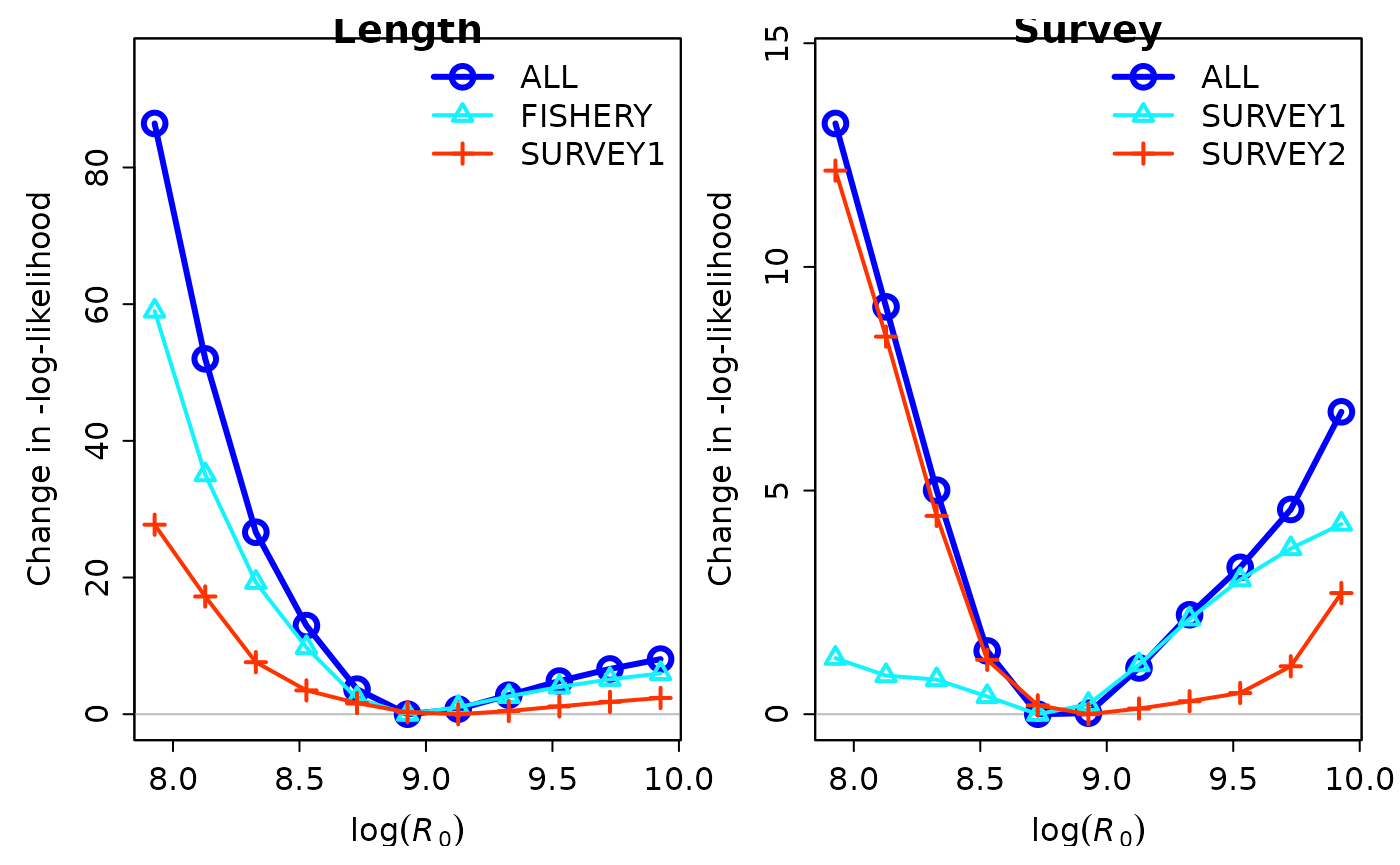
You can also plot data-type and fleet-specific profiles using
r4ss::PinerPlot(). Below we are plotting the profile for
the length composition data by fleet and the likelihood of survey data
by fleet. This will allow us to see if there are conflicts and what
sources are the main drivers.
sspar(mfrow = c(1, 2))
PinerPlot(profile_mods_sum,
component = "Length_like",
main = "Length"
)
#> Parameter matching profile.string = 'R0': 'SR_LN(R0)
#> Parameter values (after subsetting based on input 'models'): 7.9274, 8.1274, 8.3274, 8.5274, 8.7274, 8.9274, 9.1274, 9.3274, 9.5274, 9.7274, 9.9274,
#> Fleet-specific likelihoods showing max change as fraction of total change.
#> To change which components are included, change input 'minfraction'.
#> frac_change include
#> FISHERY 0.6825 TRUE
#> SURVEY1 0.3208 TRUE
#> SURVEY2 0.0000 FALSE
PinerPlot(profile_mods_sum,
component = "Surv_like",
main = "Survey"
)
#> Parameter matching profile.string = 'R0': 'SR_LN(R0)
#> Parameter values (after subsetting based on input 'models'): 7.9274, 8.1274, 8.3274, 8.5274, 8.7274, 8.9274, 9.1274, 9.3274, 9.5274, 9.7274, 9.9274,
#> Fleet-specific likelihoods showing max change as fraction of total change.
#> To change which components are included, change input 'minfraction'.
#> frac_change include
#> FISHERY 0.0000 FALSE
#> SURVEY1 0.3218 TRUE
#> SURVEY2 0.9202 TRUE